Turning Clinical Data Into Research-Driven Discoveries
i2b2 – Point-and-click interface for cohort discovery across large clinical datasets. Quickly build complex queries without needing to write code.
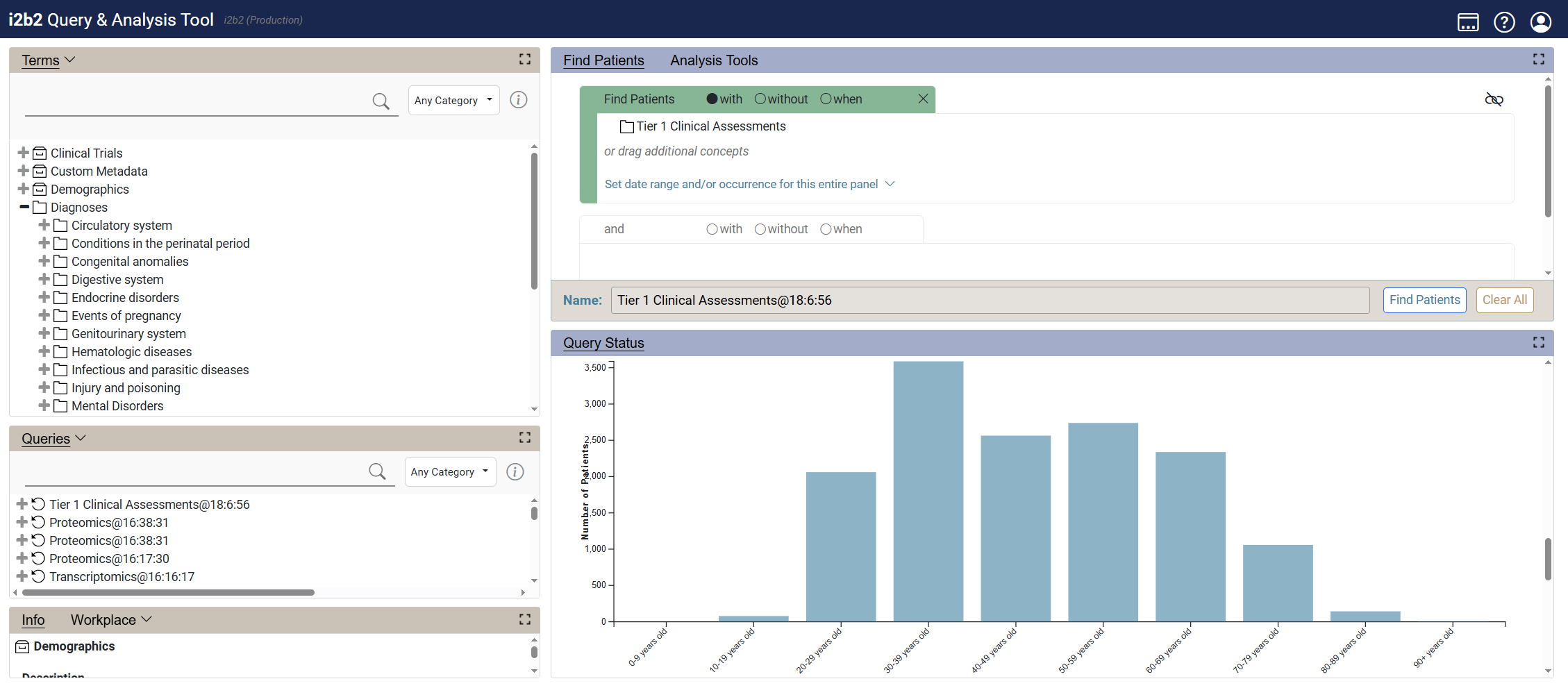

Cohort Discovery
Clinical Data
Self-service Queries
Secure Access
Interoperability
Open Source
Data Exploration
Seamless Data Integration
i2b2 connects with diverse clinical and research data sources to accelerate discovery and insight.
Data from OMOP
i2b2 supports standardized datasets from the OMOP Common Data Model, allowing researchers to leverage observational data across institutions for large-scale analytics.
Learn moreImport from REDCap
Easily load CRF (Case Report Form) data directly from your REDCap instances into i2b2, enabling streamlined research workflows and integration with other clinical datasets.
Learn moreData from EPIC
Clinical data from EPIC electronic health record (EHR) systems can be imported into i2b2, making patient demographics, encounters, diagnoses, labs, and medications available for translational research.
Learn moreOmics Data Integration
i2b2 enables researchers to bring in genomic datasets, linking molecular profiles with clinical data to support precision medicine and translational research initiatives.
Learn moreFrom Data to Discovery

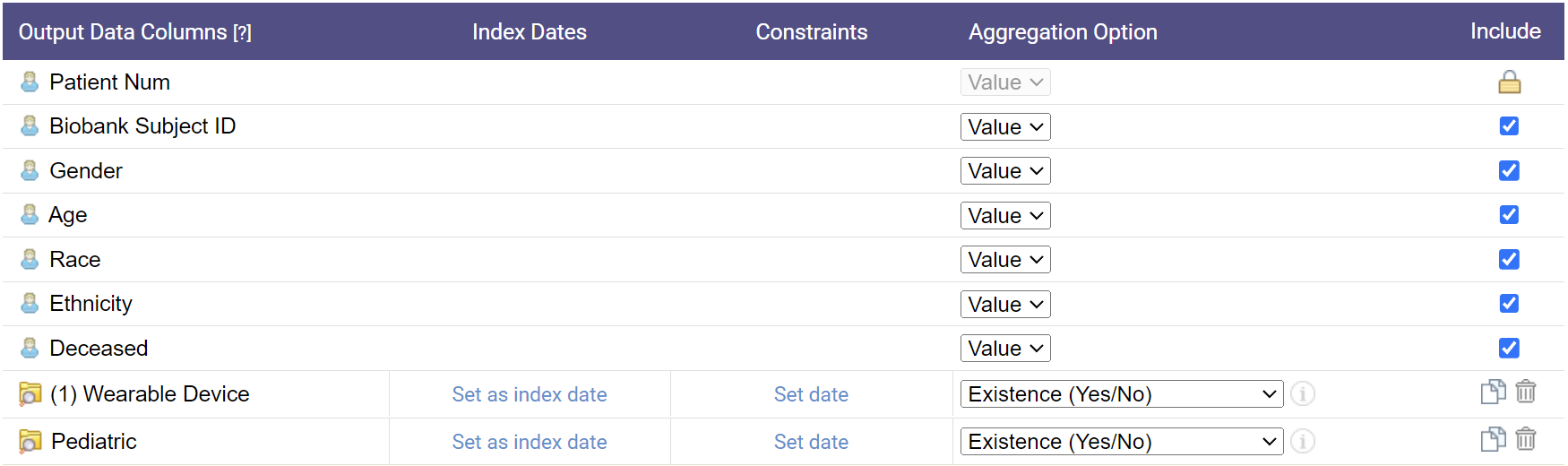
1. Finding a Patient Cohort
Identify the right group of patients for your research by combining diverse clinical and research data elements.
- Search across demographics, diagnoses, labs, medications, and more
- Refine search queries with filters and constraints
- Design complex, logical, and temporal cohort definitions
2. Exporting Aggregated or Full Datasets
Once your cohort is defined, export the data in the format that best supports your research needs.
- Generate aggregated counts and summary statistics for feasibility studies
- Export de-identified patient-level datasets for analysis
- Choose standardized formats that integrate with external tools and pipelines



3. Data Science and Analytics
Bring your data into advanced analytic environments to uncover patterns and insights.
- Integrate with R, Python, and machine learning workflows
- Perform statistical analysis, visualization, and modeling
- Enable reproducible research and support AI-driven discovery
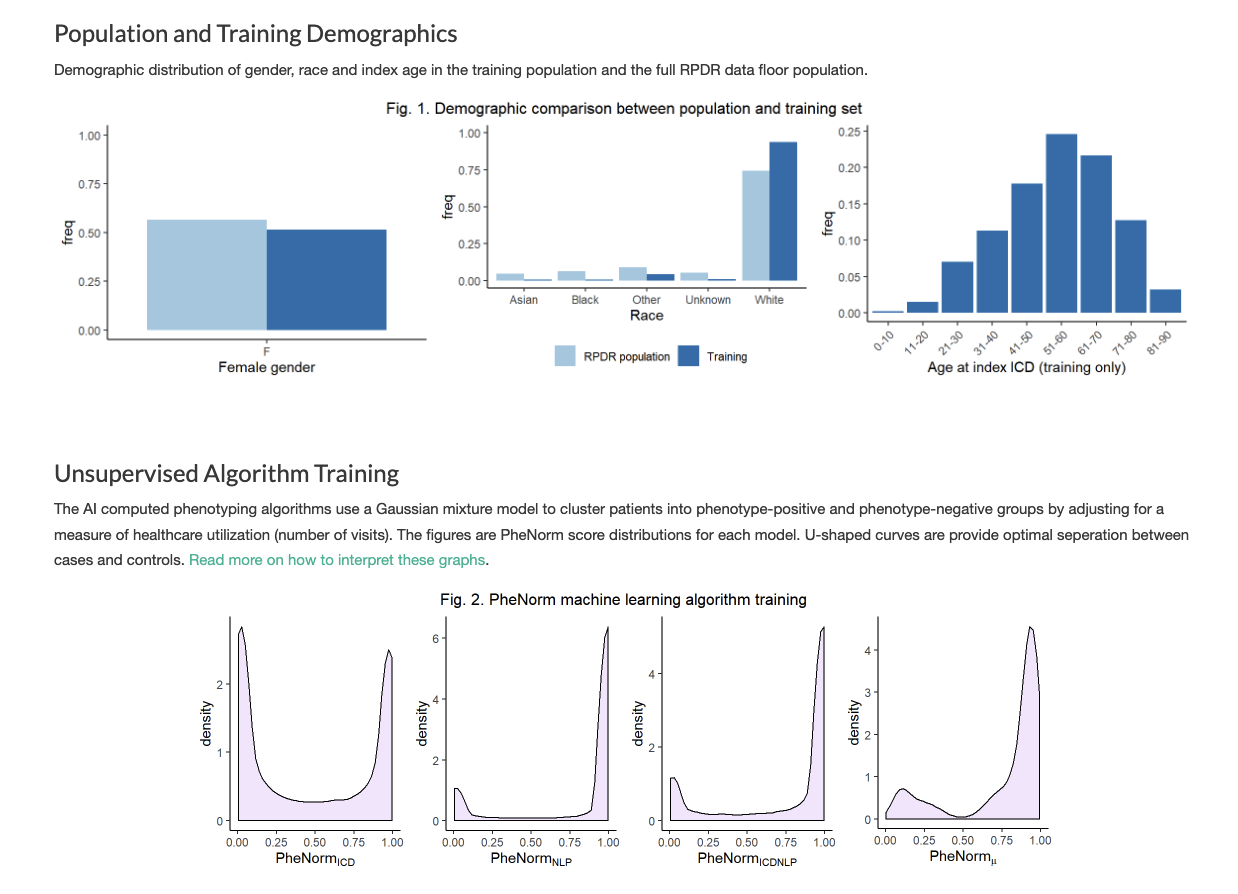
4. Computed Phenotypes
Empowering investigators to define, validate, and apply computable disease definitions and clinical characteristics at scale.
- Create and refine disease cohorts using EHR-based machine learning
- Validate and share phenotype algorithms across research groups to ensure reproducibility.
- Integrate computed phenotypes into downstream analyses, supporting clinical discovery and translational research.

Why Researchers Trust i2b2
What Researchers Are Saying






Clinical data warehouses provide harmonized access to healthcare data for medical researchers. Informatics for Integrating Biology and the Bedside (i2b2) is a well-established open-source solution with the major benefit that data representations can be tailored to support specific use cases.
BMC Medical Informatics and Decision Making volume 24, Article number: 333
2024
The i2b2 data warehousing and analytics platform is used at over 200 sites worldwide, which uses a flexible ontology-driven approach for data storage. We previously demonstrated this ontology system can drive data reconfiguration, to transform data into new formats without site-specific programming.
PLoS ONE 14(2): e0212463.
2019
Frequently asked questions
Everything you need to know about i2b2 and our software.
What is i2b2?
i2b2 (Informatics for Integrating Biology & the Bedside) is an open-source platform that allows researchers to query, explore, and analyze clinical and research data. It’s widely used for cohort discovery, feasibility studies, and translational research.
Who uses i2b2?
i2b2 is used by academic medical centers, hospitals, research institutes, and consortia worldwide. It supports clinicians, data scientists, and biomedical researchers working with large, complex datasets.
What types of data can i2b2 handle?
i2b2 supports a wide variety of data, including demographics, diagnoses, procedures, lab results, medications, clinical notes, REDCap forms, EHR data (such as EPIC), OMOP CDM data, and even genomic datasets.
How does i2b2 protect patient privacy?
i2b2 is designed to work with de-identified or limited datasets in compliance with HIPAA and institutional standards. It provides secure, role-based access and can be deployed in safe-harbor environments.
Can i2b2 integrate with other tools?
Yes. i2b2 integrates with REDCap, OMOP CDM, and major EHR systems like EPIC. Data can be exported into R, Python, or other analytics environments, making it easy to use i2b2 alongside data science and machine learning workflows.
How do I get started with i2b2?
i2b2 is open source and freely available. Institutions can deploy it locally or within their clinical data warehouse environment. Documentation, support, and an active community are available through the i2b2 website and its open-source consortium.
